Guest blog by Evangelia Michaloudi, Polyxeni Kourkoutmani and Elisavet Kaitetzidou
The use of environmental DNA (eDNA) over the past decade for monitoring aquatic biodiversity has surged due to advancements in molecular methods enabling the extraction of information from tiny traces of DNA shed by aquatic organisms into their environment. eDNA is proving to be increasingly time and cost effective for monitoring, with as little as a few hundred milliliters of filtered water being sufficient to identify everything from microscopic species of plankton up to larger organisms such as fish. eDNA is particularly useful for elusive and taxonomically complex species not easily identified through morphology alone.
By contrast, traditional methods applied to aquatic biodiversity studies require substantial volumes of water (e.g., at least 30 liters when sampling zooplankton in a lake) to be collected and filtered with time consuming microscopic analysis requiring taxonomic expertise to follow. Even then, elusive species may be missed and taxonomically challenging samples such as damaged individuals, developmental stages, and cryptic and non-indigenous species, may remain unidentified to the species level.

Figure 1: Non-indigenous species (NIS) found/recorded in Thessaloniki Bay; (a) the calanoid copepod Pseudodiaptomus marinus and (b) the cyclopoid copepod Oithona davisae; bar scale 50 μm
In the ZooPlankton Lab within the Laboratory of Ichthyology of the School of Biology in the Aristotle University of Thessaloniki, we have been monitoring the zooplankton community in Thessaloniki Bay (40°37′52.7″N; 22°56′06.1″E) since 2018. This bay is next to the port of Thessaloniki, one of the main Mediterranean ports, receiving a significant number of commercial maritime vessels that discharge their ballast tanks into the bay, thereby increasing the risk of biological invasions. We use traditional methods for biodiversity monitoring that involve deploying a plankton net to a depth of 3 m, which is then vertically hauled to filter and retain zooplankton within the water column. This sampling is typically only performed during daytime (in the morning) but we also carried out some nighttime samplings. Back in the laboratory we microscopically identify the animals (as small as 50 μm), which in many cases we need to dissect, to look at the discriminating morphological characteristic that could be a spine, a seta, cilia, appendages, etc. Through this process we have recorded so far two non-indigenous species which are also first records in the Hellenic seas. These are two tiny microscopic copepods Oithona davisae (size of about 0.5 mm) (Anadoli & Michaloudi, 2019) and Pseudodiaptomus marinus (size of about 1 mm) (Kourkoutmani & Michaloudi, 2022). The latter could have easily gone undetected because it is a species that stays in the bottom during daytime only moving up in the water column during nighttime, which is when we collected it.

Figure 2: Sampling procedure when collecting eDNA with the vampire sampler in the field
eDNA would likely have detected P. marinus even during daytime sampling, highlighting the value of this monitoring tool for revealing non-indigenous invasive species. Early detection of invasives is of great importance, since in many cases these species compete with native species and cause economic and environmental harm. eDNA monitoring can be built into early detection programs, to provide data underpinning effective control measures, to limit the spread and minimize the impacts of invasive species. The BGE (Biodiversity Genomics Europe) project has a task focusing on the sampling of ports and harbors aimed at building capacity in the early detection of marine invasives using eDNA. Sampling by both scientists and citizen scientists has been carried out across Europe following standardized protocols – the simplicity of the sample collection makes it possible for non-specialist citizens and even children to participate. Our team participated in this pan-European project and collected eDNA samples in the port of Thessaloniki, and we are going to sample other ports and harbors with the help of citizens in the future.
The analysis of the eDNA samples collected during the BGE project may reveal further invasive species beyond the two we already encountered through our traditional monitoring. More generally, it will also help to identify the full spectrum of species that constitute the indigenous zooplankton community in the sampled ports. This will include at-risk species for which distributional data are much needed to support conservation measures. Using eDNA to increase the scale and speed of research into biodiversity will help us to tackle the urgent problem of massive biodiversity loss and contribute to our still limited knowledge about the numerous species that are waiting to be described and discovered. Here the results from eDNA analysis can feed back to inform the work of ecologists and taxonomists using traditional approaches, whose knowledge and judgment remain invaluable to the conservation of marine biodiversity.
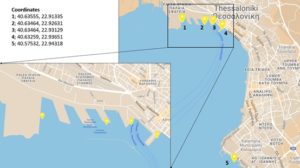
Figure 3: Location of the sampling sites in Thessaloniki Bay (northern part of Thermaikos Gulf, Aegean Sea)
References
Anadoli O & Michaloudi E (2019). Oithona davisae Ferrari & Orsi, 1984 (Copepoda: Cyclopoida: Oithonidae): Α newly recorded species in the North Aegean Sea. In: Dragicevic et al. New Mediterranean Biodiversity Records (December 2019). Mediterranean Marine Science 20 (3): 645-656. https://doi.org/10.12681/mms.20913
Kourkoutmani P & Michaloudi E (2022). First record of the calanoid copepod Pseudodiaptomus marinus Sato, 1913 in the North Aegean Sea. Thessaloniki Bay, Greece. BioInvasions Records 11: 738-746. https://doi.org/10.3391/bir.2022.11.3.15




